Projects
Here are some projects I worked on during my more academic years. Selected because I played an important role and each one of them was fun, interesting and a little different in their own ways. For a more comprehensive description of my work and academic publication check out my CV.
RppH can faithfully replace TAP to allow cloning of 5′-triphosphate carrying small RNAs
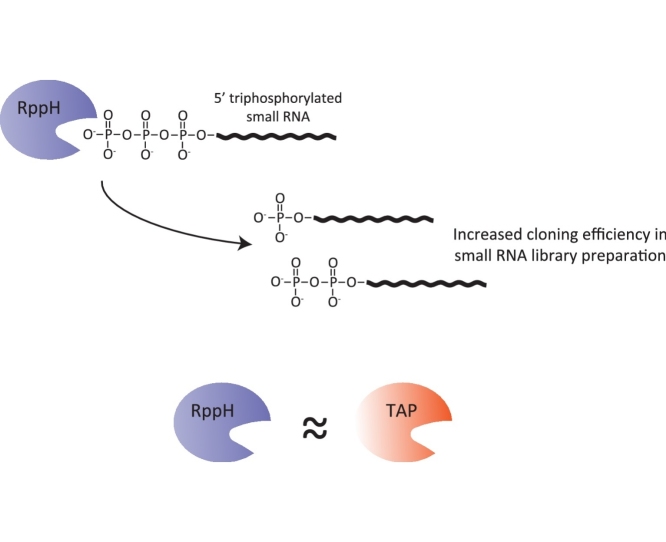
RNA interference was first described in the nematode Caenorhabditis elegans. Ever since, several new endogenous small RNA pathways have been described and characterized to different degrees. The very prominent secondary small interfering RNAs, also called 22G-RNAs, bear a 5′ triphosphate group after loading into an Argonaute protein. This creates a technical issue, since 5′PPP groups decrease cloning efficiency for small RNA sequencing. To increase cloning efficiency of these small RNA species, a common practice in the field is the treatment of RNA samples, prior to library preparation, with Tobacco Acid pyrophosphatase (TAP). Recently, TAP production and supply was discontinued, so an alternative must be devised. We turned to RNA 5′ pyrophosphohydrolase (RppH), a commercially available pyrophosphatase isolated from E. coli. Here we directly compare TAP and RppH in their use for small RNA library preparation. We show that RppH-treated samples faithfully recapitulate TAP-treated samples. Specifically, there is enrichment for 22G-RNAs and mapped small RNA reads show no small RNA transcriptome-wide differences between RppH and TAP treatment. We propose that RppH can be used as a small RNA pyrophosphatase to enrich for triphosphorylated small RNA species and show that RppH- and TAP-derived datasets can be used in direct comparison.
PERLs - PGC-expressed non-coding RNA loci
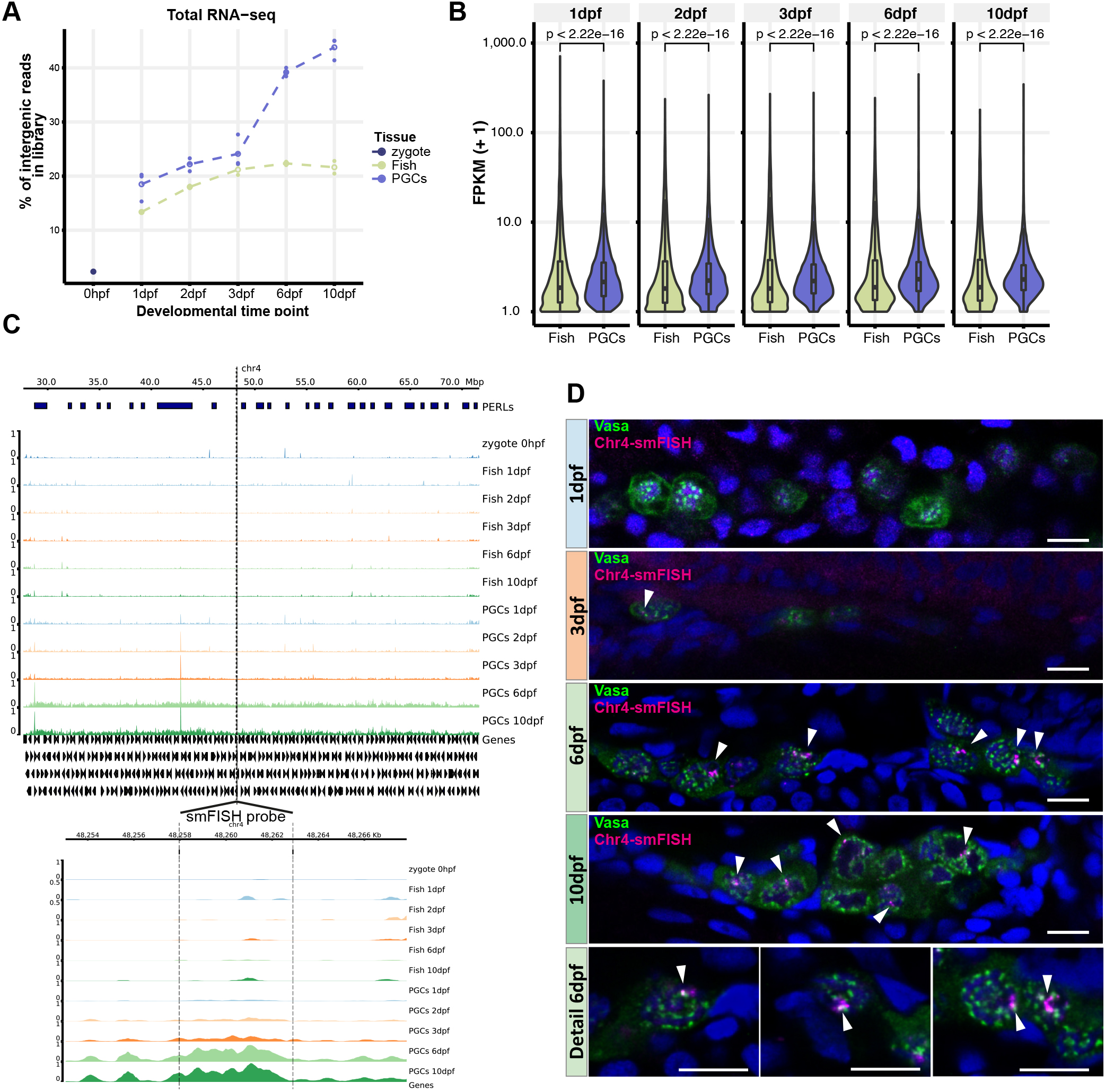
Development (2021) 148 (2): dev193060
We identify PERLs: large loci (up to several megabases in size) that are expressed from both strands in 6-10dpf PGCs. What is their function and how are they activated? Clearly, both questions are closely connected, and we cannot give definite answers. However, we hypothesize that the ‘why’ of PERLs is closely related to piRNA biology: they are rich in TEs, the piRNA coverage of PERLs is higher than that of other genomic loci and we detected PERL transcripts close to Vasa-positive granules. Furthermore, PERL transcripts start to be expressed when the zygotic piRNA machinery also starts to be transcribed, and when nuage becomes more compact, a process driven by known piRNA pathway components such as Zili and Tdrd1. Even though these are mostly correlations, and we have not yet addressed causality, these features mark PERLs as excellent piRNA cluster candidates.
Identification of Tox chromatin binding properties and downstream targets by DamID-Seq
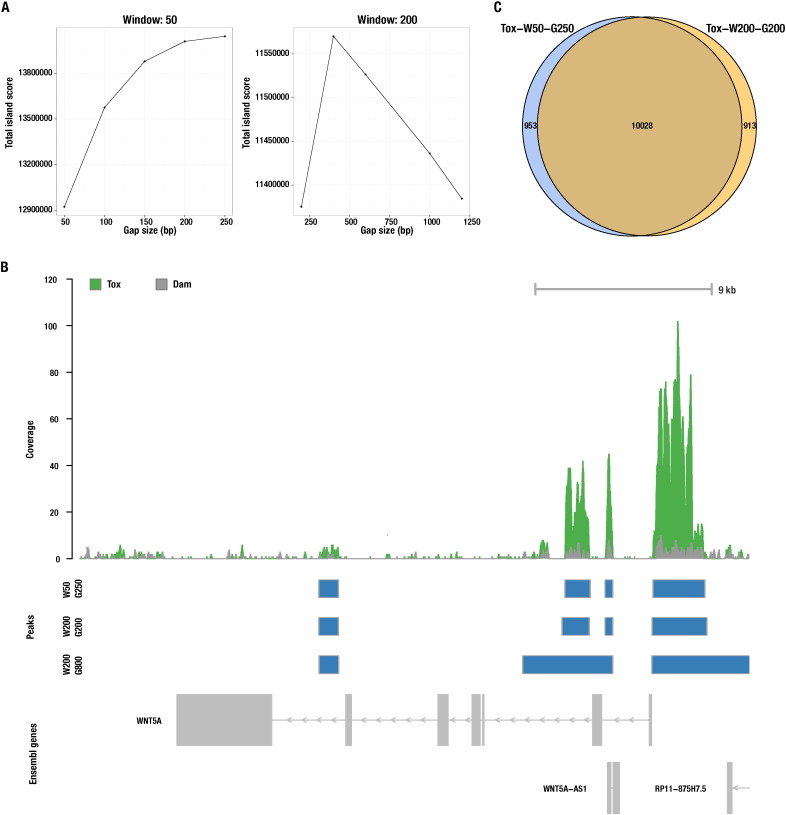
In recent years, DNA adenine methyltransferase identification (DamID) has emerged as a powerful tool to profile protein-DNA interaction on a genome-wide scale. While DamID has been primarily combined with microarray analyses, which limits the spatial resolution and full potential of this technique, our group was the first to combine DamID with sequencing (DamID-Seq) for characterizing the binding loci and properties of a transcription factor (Tox) (sequencing data available at NCBI’s Gene Expression Omnibus under the accession number GSE64240). Our approach was based on the combination and optimization of several bioinformatics tools that are here described in detail. Analysis of Tox proximity to transcriptional start sites, profiling on enhancers and binding motif has allowed us to identify this transcription factor as an important new regulator of neural stem cells differentiation and newborn neurons maturation during mouse cortical development. Here we provide a valuable resource to study the role of Tox as a novel key determinant of mammalian somatic stem cells during development of the nervous and lymphatic system, in which this factor is known to be active, and describe a useful pipeline to perform DamID-Seq analyses for any other transcription factor.
Characterization of genetic loss-of-function of Fus in zebrafish
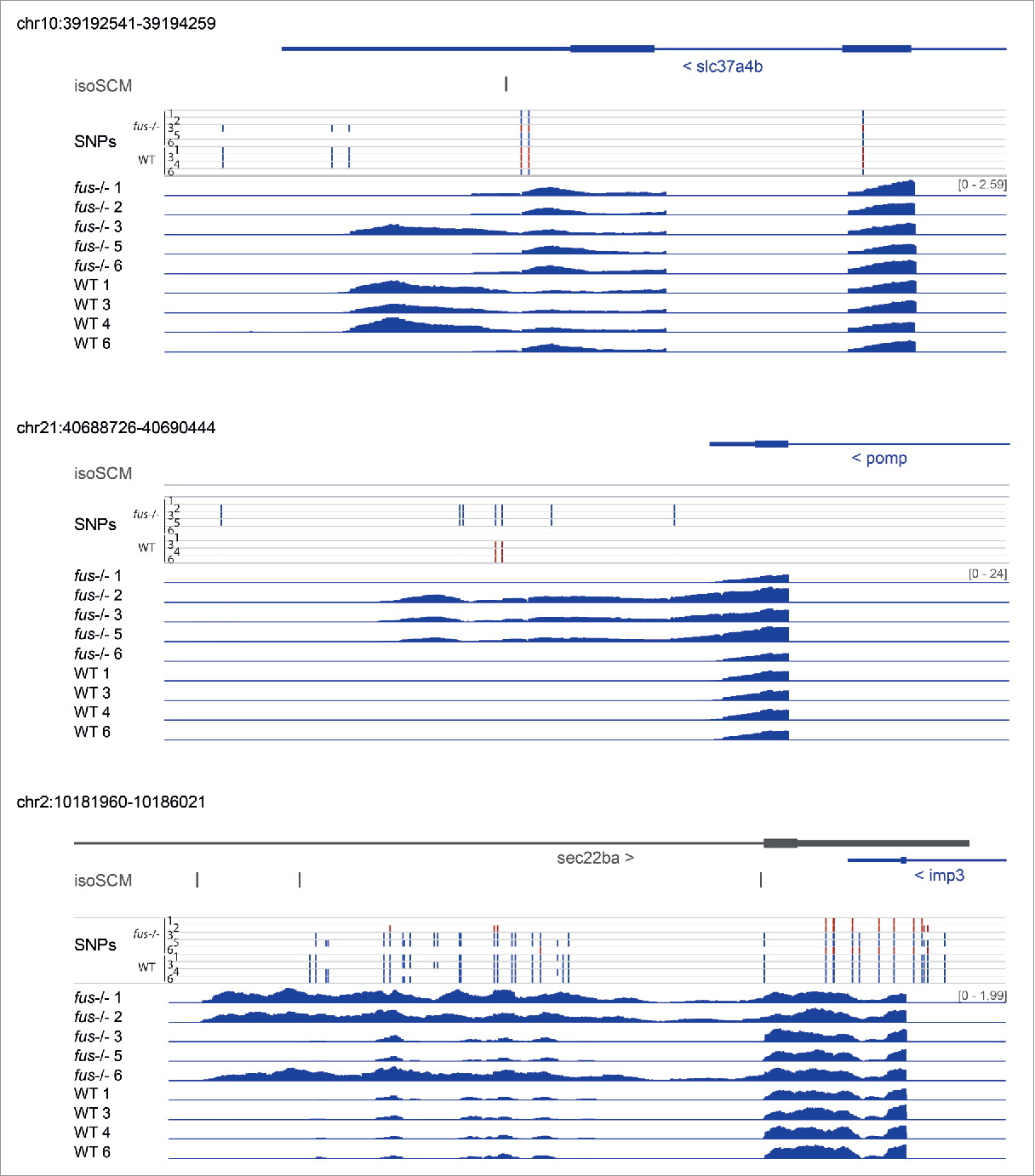
The RNA-binding protein FUS is implicated in transcription, alternative splicing of neuronal genes and DNA repair. Mutations in FUS have been linked to human neurodegenerative diseases such as ALS (amyotrophic lateral sclerosis). We genetically disrupted fus in zebrafish (Danio rerio) using the CRISPR-Cas9 system. The fus knockout animals are fertile and did not show any distinctive phenotype. Mutation of fus induces mild changes in gene expression on the transcriptome and proteome level in the adult brain. We observed a significant influence of genetic background on gene expression and 3′UTR usage, which could mask the effects of loss of Fus. Unlike published fus morphants, maternal zygotic fus mutants do not show motoneuronal degeneration and exhibit normal locomotor activity.